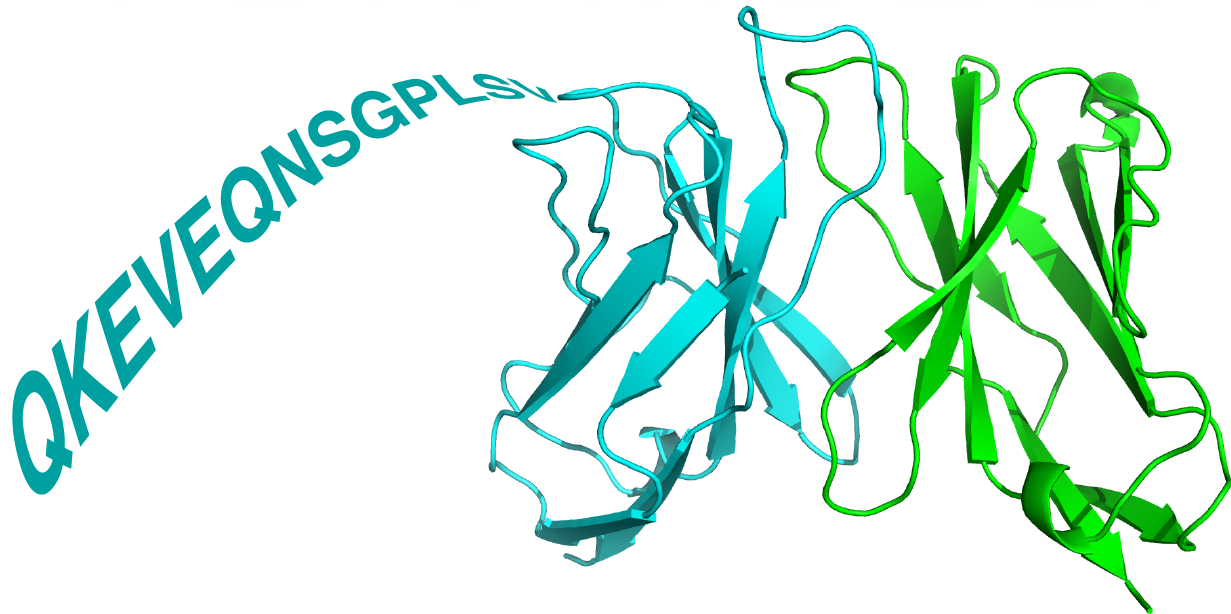
TCRmodel is a web server that models T cell receptor (TCR) structures and T cell receptor-peptide MHC (TCRpMHC) complex structures from sequence, developed by the Pierce Lab at the University of Maryland Institute for Bioscience and Biotechnology Research. This website is free and open to all users and there is no login requirement.
T cell receptors, or TCRs, are immunoglobulin immune receptors found on T cells, and each human is estimated to possess millions of unique TCRs. As with antibodies, TCRs are heterodimers, and diversity is primarily exhibited in loops called complementarity determining region (CDR) loops, which engage foreign antigens. In the case of TCRs, these are typically peptides that are presented by major histocompatibility complex (MHC) proteins. TCRs represent a key element of the cellular immune response, and have major implications in vaccine design, autoimmune disease, and as immunotherapeutics for cancer.
To address the growing need to gain structural insights of TCRs based on their sequences, due in part to advances in immune repertoire sequencing and massive increases in TCR sequence data, and fueled by the recent breakthroughs in deep learning and artificial intelligence, we released TCRmodel 2.0 and provide an easy-to-use interface for users to generate TCR or TCRpMHC models accurately and quickly, with the capability to visualize structural information in the browser. TCRmodel 2.0 leverages template-based and deep learning methods to reliably predict the structure of TCRs and TCRs in complex with their peptide-MHC (pMHC) targets.
If you used our tools, please cite:
Previously, this site performs template-based modeling of TCRs, using all known TCR structures as templates and with optimizations to correctly model TCR architectures. While we no longer support the template-based modeling protocol on this site, you may access that algorithm via RosettaTCR, in the powerful protein modeling program Rosetta. If you used the earlier version of this site or RosettaTCR, please cite:
T cell receptors, or TCRs, are immunoglobulin immune receptors found on T cells, and each human is estimated to possess millions of unique TCRs. As with antibodies, TCRs are heterodimers, and diversity is primarily exhibited in loops called complementarity determining region (CDR) loops, which engage foreign antigens. In the case of TCRs, these are typically peptides that are presented by major histocompatibility complex (MHC) proteins. TCRs represent a key element of the cellular immune response, and have major implications in vaccine design, autoimmune disease, and as immunotherapeutics for cancer.
To address the growing need to gain structural insights of TCRs based on their sequences, due in part to advances in immune repertoire sequencing and massive increases in TCR sequence data, and fueled by the recent breakthroughs in deep learning and artificial intelligence, we released TCRmodel 2.0 and provide an easy-to-use interface for users to generate TCR or TCRpMHC models accurately and quickly, with the capability to visualize structural information in the browser. TCRmodel 2.0 leverages template-based and deep learning methods to reliably predict the structure of TCRs and TCRs in complex with their peptide-MHC (pMHC) targets.
If you used our tools, please cite:
Yin R, Ribeiro-Filho HV, Lin V, Gowthaman R, Cheung M, Pierce BG. (2023) TCRmodel2: high-resolution modeling of T cell receptor recognition using deep learning. Nucleic Acids Res. In press. https://doi.org/10.1093/nar/gkad356
Pubmed
Previously, this site performs template-based modeling of TCRs, using all known TCR structures as templates and with optimizations to correctly model TCR architectures. While we no longer support the template-based modeling protocol on this site, you may access that algorithm via RosettaTCR, in the powerful protein modeling program Rosetta. If you used the earlier version of this site or RosettaTCR, please cite:
Gowthaman R, Pierce BG. (2018) TCRmodel: high resolution modeling of T cell receptors from sequence. Nucleic Acids Research, 46(Web Server issue), W396–W401. http://doi.org/10.1093/nar/gky432
Pubmed